The availability of huge omics data from genome projects and high-throughput technology (next-generation sequencing and microarray) has brought a great challenge to understand the complexity of biological processes and disease mechanisms in eye research. We seek an agile and predictive understanding of how genetic variants that results in eye diseases, including ocular cancer. We write algorithms and pipelines to get critical answers faster from NGS data. We also focus on non-coding RNA expression and its regulatory role in eye diseases by integrating data from NGS and the public. We have a reliable infrastructure and framework comprised of LINUX and Windows-based servers and desktop workstations, which allow us to integrate high throughput data and study them at the systems level. Bioinformatics centre is highly interdisciplinary, at the interface of Biology, computational biology and Informatics.
We primarily focus on developing bioinformatics methods of NGSomics data analysis and the role of small noncoding RNAs as biomarkers for vision biology and eye diseases. This includes whole genome, exome, transcriptome next-generation sequencing data analysis in identifying molecular targets for ocular cancer and diseases, the small-RNA seq analysis of human with focus on profiling micro-RNAs and identifying their regulatory role in eye diseases, comparative genome analysis to identify virulence and drug resistance mechanisms in ocular pathogens and structure-based approaches of non-synonymous variants (nsSNVs) to understand the molecular mechanisms for pathogenicity. As part of the multidisciplinary nature of our field, we work on these projects in close collaboration with wet-lab scientists. The following gives a short summary of a few selected research projects.
Genomics of Eye Diseases:
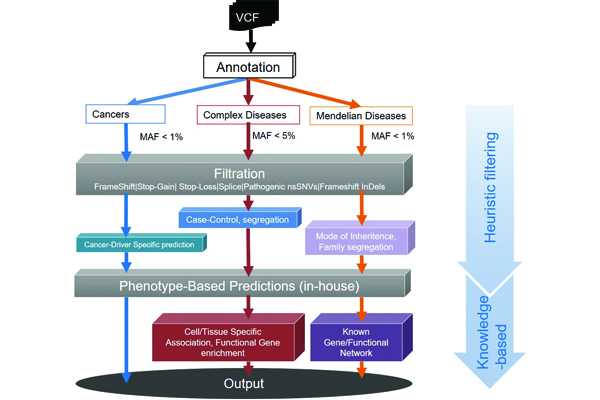
Recent advances in next generation sequencing (NGS) methods have brought a paradigm shift in discovering eye disease-associated genetic variants from linkage and genome-wide association studies to NGS-based genome/exome studies. Mainly, whole-exome sequencing (WES) is now made as a viable approach to uncover the pathogenic variants for both Mendelian and complex eye diseases with a limited number of probands. WES, focuses on only the protein-coding sequence of the human genome, is become a powerful tool with many advantages in the research setting, and moreover, is now being implemented into the clinical diagnostic arena. Nevertheless, the identification of pathogenic variants remains a great challenge. Pathogenic variant prioritization using simple heuristic filtering approaches and functional implications of variants misses the true positives. Using our in-house automated pipeline, we can find almost all variants within the targeted region of the genomes. We use stringent filtering methods and machine learning methods to prioritize pathogenic variants for eye diseases. We further aim to achieve a variant prioritized model to correctly filter eye disease-causing variants for Mendelian and complex eye diseases. In tandem, we work on translational genomics of ocular cancer including Retinoblastoma and ocular lymphoma.
Comparative Genomics of ocular bacterial pathogens:
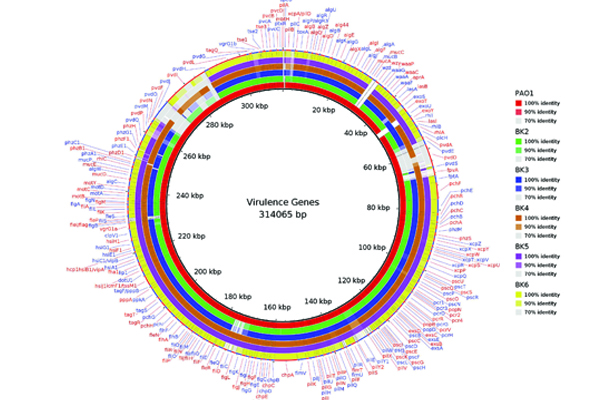
Microbial keratitis due to either fungus or bacteria is a major cause of blindness in India. The bacterial keratitis, often caused by Pseudomonas aeruginosa and methicillin-resistant staphylococcus aureus (MRSA) at Aravind Eye hospital, Madurai, majority of times in spite of adequate medical management the ulcer does not heal and may require a corneal transplant. Pseudomonas aeruginosa can cause a most severe keratitis, carrying a wide array of virulence factors that contribute pathogenesis. Keratitis pathogenesis is a complex process, where in several virulence factors has been implicated including Cell-associated structures such as type IV pili and flagella, slime polysaccharide, proteases such as elastase B (LasB), alkaline protease, protease IV and Pasp and exotoxins. Also, clinical isolates of Pseudomonas often exhibit multiple resistances to antibiotics. On the other hand, studies have shown an increase in the pervasiveness of ocular MRSA infections. Comparative genomics through de novo assembly of next-generation sequencing and third-generation sequencing methods of ocular bacterial pathogens show specific genetic suggesting that to cause eye infections. Further insights show that these strain-specific features may response to the host immune system, which indeed affects the outcome of the disease.
Human microRNAs and their regulatory role in eye diseases
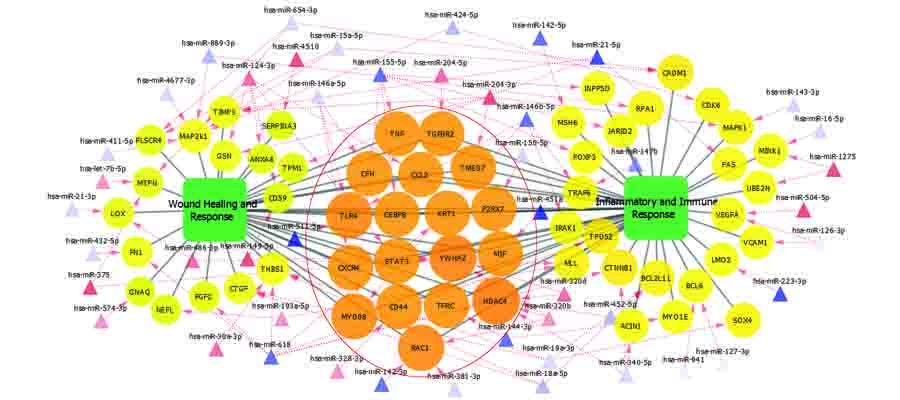
MicroRNAs are a novel group of non-coding small RNAs that post-transcriptionally control gene expression by promoting either degradation or translational repression of target messenger RNA. They are implicated in a large variety of physiological and pathophysiological processes. Levels of miRNAs in the serum of humans have been shown to be stable, reproducible, consistent amongst healthy individuals and change during pathophysiology, and their presence in ocular fluids allowing them to be of potential value as clinical biomarkers of eye disease. We profile miRNAs using deep sequencing methods and predict their regulatory role in ocular tuberculosis, glaucoma, diabetic retinopathy and microbial keratitis.
Structural Bioinformatics of eye disease-associated single nucleotide variations
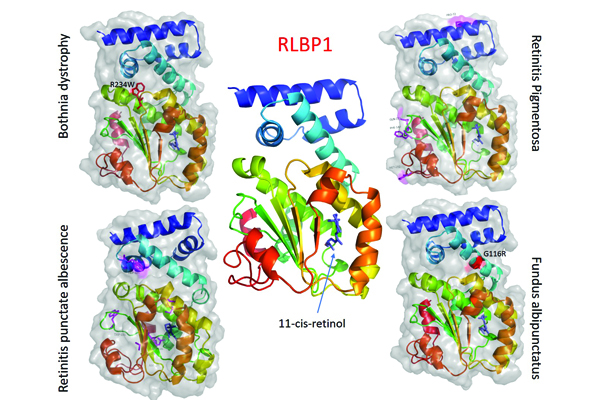
The advent of NGS identified several new candidate genes for eye diseases. Variations in the same gene can cause very different eye diseases (pleiotropy), on the other hand, the single disease can be genetically heterogeneous. Thus, a detailed underlying molecular mechanism is needed to understand this complexity. Non-synonymous single nucleotide variants (nsSNVs) in the coding region of the protein is of critical importance to understand the molecular mechanisms of the disease and to clarify the association between patient-specific variants and disease phenotype. We use simple analytical strategy using protein structure to predict the pathogenicity of nsSNVs and investigate their functional impact that may lead to eye disease phenotypes.
Scientist
- Dr. Bharanidharan Devarajan
Research Scholars
- Mr. K. Manoj Kumar
- Mr. K. Kathirvel
- Ms. Swathi CH
- Mr. A.Mohamed Hameed Aslam
- Ms. O. Ruthra
AMRF Biocomputing Center (ABC)
The AMRF Biocomputing Center (ABC) provides a core computational facility with a reliable infrastructure and framework equipped with Dell T630 Server (With Ubuntu 14.04) and HP DL580R07 (E7) CTO Server, three Dell workstations and five Intel i7-3370 3.5GHz workstations. It is a multidisciplinary research environment that provides to customize data analysis tailored to the needs of individual research projects across all the research groups and extend this service to others on mutually acceptable terms. In addition, it helps to train manpower by way of workshops and short training courses. AMRF, Science and Engineering Research Board (SERB) and Department of Biotechnology (DBT) Govt. of India supported for the funding.
Next-generation sequence data processing and analysis: The existing facilities has power to handle human whole genome/exome, transcriptome sequencing data. Automated Clinical Whole/exome pipelines have been developed for Mendelian and Complex Disorders. Besides, we provide data analysis support for deep-sequencing data of Small-Noncoding RNAs including microRNA and PIWI-RNAs, metagenomics, comparative genomics of microbial pathogens using both second and third generation sequencing data. The input to the NGS pipelines is either raw reads from the sequencing machines or mapped reads from alignment software.