We provides a thriving environment for basic, clinical and translational research.

Dr. Sundaresan’s team primarily focuses on understanding the molecular genetics and allied mechanisms of various ocular disorders by identifying and characterizing the candidate genes, their mutation spectrum and other possible risk factors for early intervention, wherever possible. Based on this, established genotype-phenotype correlations could be useful for specific eye disease prognosis. This arm of AMRF employs various methodologies including molecular genetics, molecular biology, next-generation sequencing technologies to answer these questions and gain insights into genetic basis, disease pathogenesis and its progression.

Dr. Vanniarajan’s team focuses on developing diagnostic, prognostic and theranostic tools for ocular tumors. This team works closely with clinicians to solve major problems such as development, progression and treatment resistance in ocular tumors and help them to improve the disease prognosis. Cutting-edge technologies such as Massive Parallel Sequencing, Bead Array, In vitro functional assays are used to study genomic, epigenomic and cellular alterations. The lab has developed a unique strategy for rapid and efficient genetic testing of retinoblastoma and successfully translated for clinic testing of RB1.
Increased burden of inherited eye diseases in India is due to endogamous populations. Approximately 4000 diseases/syndromes affect humans, among them one third is related to ocular disorders. Therefore, it is vital to improvise our knowledge along with sincere efforts towards research thus contributing to reduce disease burden. Our research group focusses on several aspects including identifying disease-causing gene/ associated genes and their mutation spectrum as well as understanding the molecular mechanisms responsible for various eye diseases such as cataract, glaucoma, diabetic retinopathy, Globe anomalies, pseudoexfoliation syndrome, Congenital Hereditary Endothelial Dystrophy, Leber’s hereditary optic neuropathy, Leber’s congenital amaurosis, Retinoschisis and other retinal dystrophies.
Glaucoma:
Glaucoma represents the second leading cause of irreversible blindness worldwide, affecting more than 12 million people in India. It is a neurodegenerative disorder characterized by irreversible damage to the optic nerve, loss of retinal ganglion cells (RGCs) and eventually ends to gradual loss of visual field, thus affecting quality of life. Actual mechanism of glaucoma is complex due to various pathways involved in its pathogenesis. Over the past two decade, our department has been involved to understand the glaucoma pathogenesis by employing basic, molecular techniques and gene expression analysis.
Outcome of our research findings including, screened several candidate genes of Primary Open Angle Glaucoma – myocilin (MYOC), CYP1B1, LTBP2, SIX6, MMP9 for mutations in South Indian families. Collectively, these studies provide insights and expand the mutational spectrum of glaucoma candidate genes in the Indian population. Further, zebrafish transgenesis experiments and luciferase assays were employed to assess the impact of a novel 4 bp deletion in the SIX6 gene, which clearly demonstrated the significantly reduced reporter expression in cells of the retinal ganglion and amacrine layers, where human SIX6 is expressed (Mol Genet Genomic Med. 2017).
Furthermore, in a collaborative genome wide association study (GWAS), 5 new susceptibility loci for primary angle closure glaucoma was identified, with PCMTDI-ST18 (SNP rs1015213) found to be the most significant one in south Indian patients (Nat Genet, 2016; IOVS, 2013). We have also explored the association of LOXL1, CACNA1A, CLU and TNF-α, MTHFR genes to understand their role in south Indian PEX patients (Nat Genet. 2015; Curr Eye Res. 2015; Nat Genet. 2017; Hum Mol Genet. 2019). Besides, investigation of aqueous cytokines in angle closure glaucoma patients revealed significantly higher levels of IL-8, eotaxin, IP-10 and MIP-1β in the chronic PACG eyes suggesting their plausible role in PACG pathogenesis (Current Eye Research, 2017).
Diabetic Retinopathy:
Diabetic retinopathy (DR) is the leading cause of visual disability associated with microvascular changes in the retina. According to the WHO reports, India will become the major hubs of diabetic population during the next 2 decades. As genetic factors known to play crucial role in DR onset, molecular genetic studies contributed significantly towards the understanding of DR pathogenesis.
Our research team has extensively worked in the past years to ascertain the genetics of DR. Several polymorphic variants of candidate genes including VEGF, eNOS, RAGE, PEDF, AKR1B1, EPO, HTRA1, ICAM, HFE, CFH, and ARMS2 screened in South Indian DR patients, reported the significant association of SNP in VEGF and RAGE with DR associated mechanisms (Mol Vis, 2006, BMC Med Genet, 2010). Concurrently, in a collaborative work, for the first time, we reported the association of 17q25.1 locus (SNP rs9896052) with diabetic retinopathy through a GWAS (Diabetologia, 2015). Additionally, our efforts also contributed towards the establishment of retinal mitoscriptome gene expression signatures using customized human mitochondria-focused gene chip to identify genetic markers in health and disease condition (Mitochondrion, 2017).
Oculocutaneous Albinism:
Genetic screening was established for all the candidate genes of OCA and OA, including TYR, P and MC1R which helps us to identify the risk groups and forewarning them by counseling to prevent further occurrence of the disease and facilitate prenatal diagnosis.
Congenital Hereditary Endothelial Dystrophy:
Congenital hereditary endothelial dystrophy (CHED) is a rare and bilateral congenital disorder, characterized by diffuse clouding of corneas. Though the incidence of CHED is quite low, it is more prevalent in the regions practicing consanguineous marriages like India. Our research groups have also contributed for the identification of the first candidate gene, SLC4A11 underlying CHED2. This research work provides a valuable tool for studying the gene regulation events in CHED pathology (Nature Genetics 2006).
Translational Genomic Research in Progress for Paediatric eye diseases:
Leber’s congenital amaurosis:
Leber’s congenital amaurosis (LCA) is the severe form inherited retinal dystrophy (IRD) resulting in associated vision loss at an early age. LCA displays a wide genetic and phenotypic heterogeneity with several overlapping clinical features with other forms of retinal dystrophies, thus marking the importance of genetic testing. To address this, we do mutation screening for LCA associated genes and understanding the spectrum of mutations, their effects to gain insights in its pathogenesis.
Leber’s Hereditary Optic Neuropathy:
Leber’s Hereditary Optic Neuropathy (LHON) is an inherited mitochondrial disorder, characterized by bilateral, painless subacute loss of central vision that preferentially affects young males. Unlike western population, reduced frequency of three primary mitochondrial mutations was reported in Indian population (Ophthalmic genetics, 2010). Our research focusses on screening the whole mitochondrial genome for LHON patients and evaluates the role of mitochondrial DNA variants, their prevalence and mitochondrial haplogroups. Besides, we are also focusing on understanding the intricacies of mitochondrial and nuclear genome cross-talk which will shed light in precise diagnosis of mitochondrial diseases involving ocular manifestations.
Juvenile-X-linked retinoschisis (JXLR):
Juvenile-X-linked retinoschisis (JXLR) is the leading cause of macular degeneration in males, caused by mutations in the RS1 gene. Our focus is on screening for RS1 gene mutations and evaluating the associated clinical parameters for understanding the genotype-phenotype correlations.
Molecular Analysis of Ocular Cancers
Comprehensive Genetic Analysis of Retinoblastoma
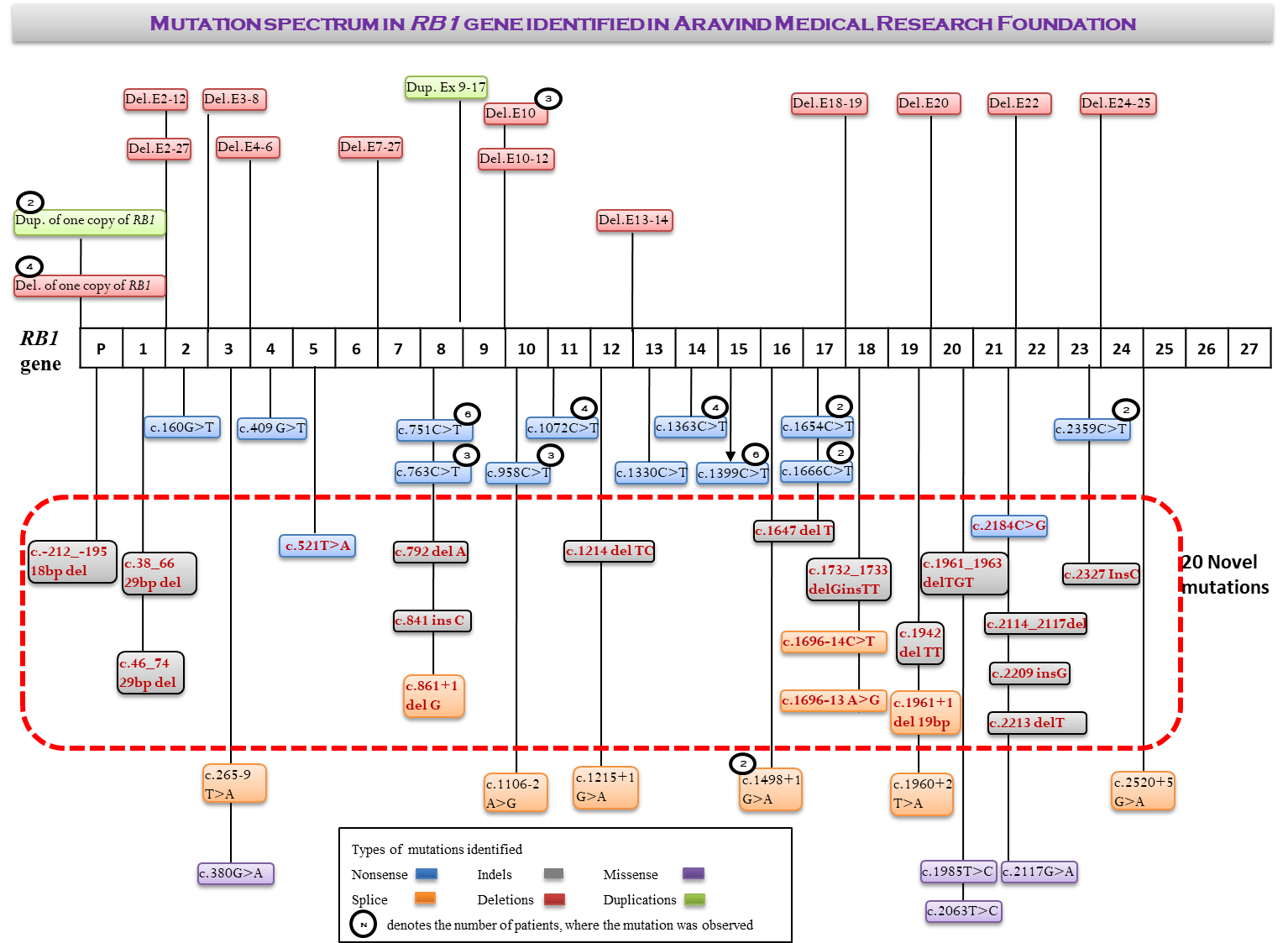
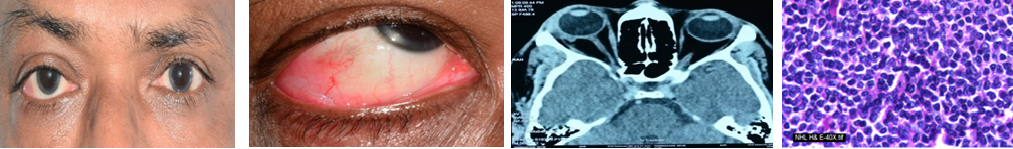
The major focus of our research work is to develop methods for comprehensive genetic analysis of retinoblastoma. RB1 is a large gene, spanning around 180 kb with 27 exons. In order to cover the entire gene, we employed Sanger sequencing, Multiplex Ligation Dependent Probe Amplification and Next Generation Sequencing. Since this involves lot of cost and time, we established a cost-effective and time-efficient strategy based on mutational frequencies of each exon. The genetic analysis is performed in tumour from enucleated globes along with blood samples of patient and family members. Segregation pattern of the mutations are further evaluated and risk is predicted and genetic counselling is provided accordingly. We have achieved 90% sensitivity of genetic testing for germline mutation in bilateral and somatic mutation in unilateral RB patients. This project is funded by Aravind Eye Foundation, USA. Based on the methods developed in this project, Clinical Genetic Testing is offered through Cancer Genetic Testing Centre of Aravind Eye Hospital, Madurai.
Analysis of RB1 transcriptional alterations
Mutational spectrum varies widely from a point mutation to the deletion of whole RB1 gene. Functionally, it includes Nonsense, Missense, Frameshift and Splice variants. Nonsense mutations are highly penetrant compared to other mutation types. Splice variants may cause altered splicing by exon skipping or intron retention. Hence the mutational effect is studied at transcript level to understand its significance. In addition, we have also established method for genetic analysis of RB1 gene from FFPE tissues. This project is funded by Indian Council of Medical Research.
Understanding key modifier genes of RB tumorigenesis
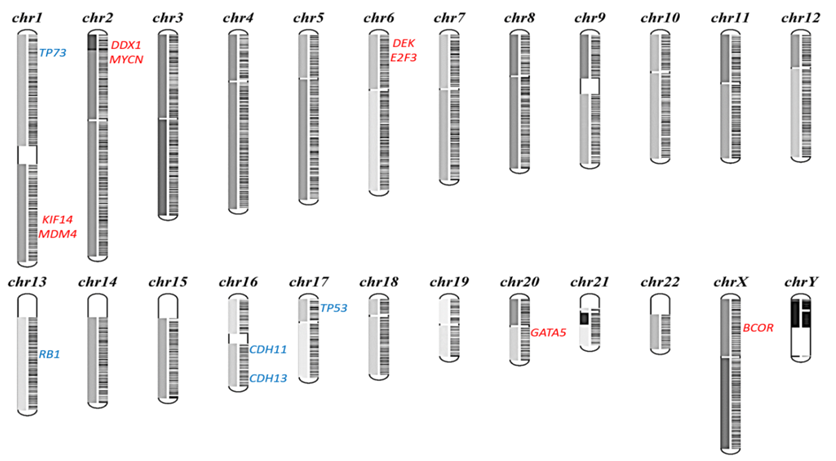
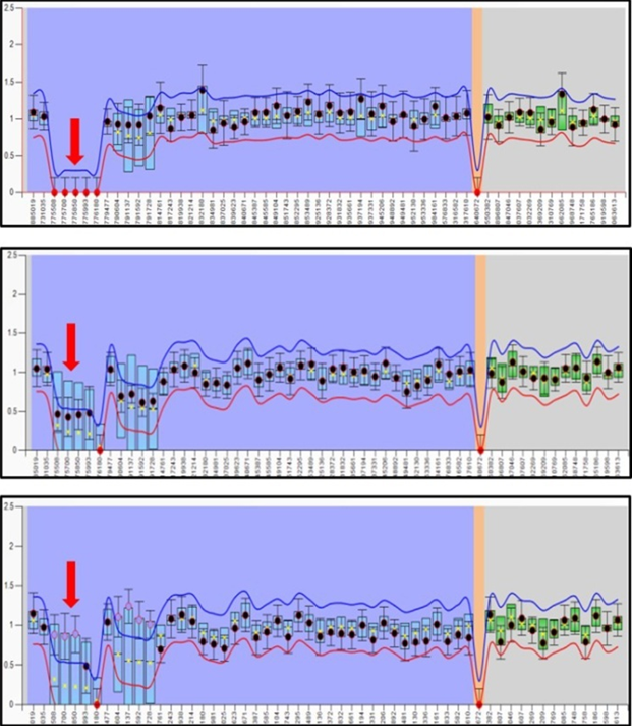
Bi-allelic inactivation of RB1 gene is required for the initiation of retinoblastoma. However, the mechanisms that enable retinoblastoma cells to acquire the additional hallmarks of cancer remain elusive. Recent research had suggested that additional genetic alterations other than RB1 inactivation are involved in the process of RB tumorigenesis. We have investigated the status of potential modifier genes in regions of recurrent gains or losses within RB genome. Targeted Next Generation sequencing of 17 genes showed significant number of copy number gains and losses of oncogenes and tumor suppressor genes respectively. Microsatellite regions in these regions showed significant correlation with the genomic instabilities in some tumors. This study not only aides in expanding our understanding of the biology of retinoblastoma but also point out immediate therapeutic targets. This project is supported by Aravind Eye Foundation, USA and Department of Biotechnology, India.
Elucidating the epigenetic events in RB tumour progression
Tumour progression in RB has been recently postulated to be driven by epigenetic mechanisms more than mere alterations of the genome. It is shown that inactivation of RB1 deregulates multiple oncogenic and tumor suppressor pathways through epigenetic mechanisms. In cancers, epigenetic mechanisms such as promoter methylation, histone modification and miRNA regulation were reported. We are looking at the promoter methylation using Methylation specific MLPA (MS-MLPA) and Bisulfite sequencing. We have also adapted genome wide methylation studies such as Infinium bead methyl array and found significant alterations. We are investigating the epigenetic changes during the progression of tumor from retinoma to retinoblastoma. We also analysed RB tumor samples for the presence of HPV genome which was reported to be cause of RB in few studies. This project is funded by Department of Biotechnology and supported by DST-INSPIRE Research fellowship.
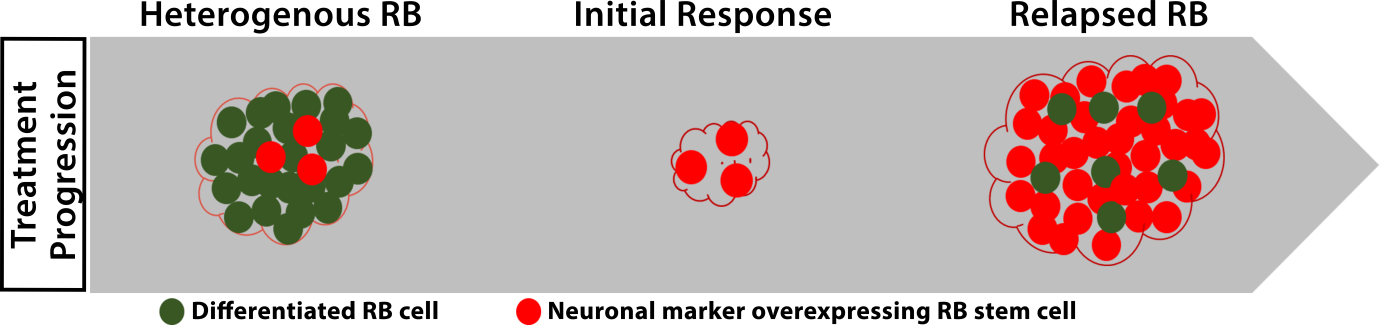
Evaluating the mechanism of chemoresistance in RB
The delayed presentation of RB leads to advance stage of the disease in Indian Scenario. Chemotherapy remains as a major mode of treatment in the advanced stage at least to save one eye in bilateral patients. However, the treatment failure is noted in about 30% due to chemoresistance. Chemoresistance is clinically defined as either a lack of reduction in tumor size or recurrence of tumor following chemotherapy. Chemoresistance may be inherent or acquired during the course of treatment. We have evaluated the expression of ATP Binding Cassette (ABC) transporters that are drug efflux pumps. We also identified the potential role of the stem cell markers in causing the chemoresistance. These cancer stem cells remain quiescent during the drug treatment and proliferate further when the drug is stopped. We are further characterizing the functional alterations in chemoresistant tumors. This project is supported by Department of Biotechnology and CSIR Research Fellowship.
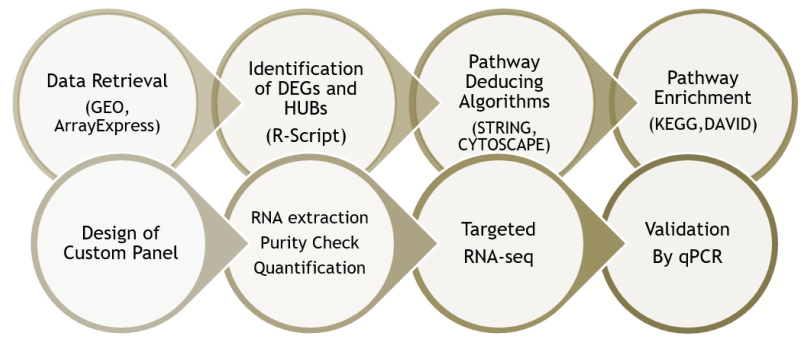
Deducing deregulated cancer related pathways in RB
Gene expression studies are the keys to conclude any genetic alterations at the DNA level changes. It is possible to deduce the pathways by compiling the changes in the regulatory genes. The understanding of the interaction of the differentially regulated genes with RB1 could provide the mechanism involved in tumorigenesis. Earlier studies by microarray had suggested that PI3K-AKT-mTOR pathway genes were predominantly expressed in Rb tumors compared to normal. Other studies with matched normal and tumor from the same individual, showed variable expression of genes related to DNA damage response. We have carried out the computational analysis with the publicly available datasets and identified the key genes. Further to validate those genes and deduce the pathway, we performed the analysis of the PCR array for the cancer pathways. This project is funded by Science and Engineering Research Board, India.
Molecular Pathological analysis of ocular cancers
Ocular cancers are unique among the other diseases of the eye, threatening both vision and life. The most common cancers of eye in elderly patients include Carcinoma, Conjunctival neoplasia, Choroidal melanoma and Ocular adnexal lymphoma. Lymphoma is the neoplastic proliferation of lymphocytes at various stages of differentiation. Ocular lymphomas are largely Non-Hodgkin’s type and of B- cell origin (about 85%). Ocular lymphoma can either present in intraocular or ocular adnexal structure. Currently diagnosis of ocular lymphoa is made with clinical and histological investigations. Few studies have indicated the importance of Immuno histochemical subtyping of tumour cells based on their origin in predicting the disease survival and prognosis in OL. Molecular diagnosis along with the subtyping of cell type would be helpful in determining suitable therapy. This project is supported by Aravind Eye Foundation, USA.
Faculty
- Dr. P. Sundaresan, Scientist
- Dr. A. Vanniarajan, Scientist
Research Associate
- Dr. Roopam Duvesh
Research Scholars
- Mrs. M. Durga
- Mr. R. Kadarkarai Raj
- Ms. P. Gowri
- Mr. C. Prakash
- Ms. A.S. Sriee Viswarubhiny
- Ms. Susmita Choudhary
- Mr. K. Thirumalairaj
- Mr. A. Aloysius Abraham
- Mr. T. S. Balaji
- Ms.T. Shanthini
- Mr. K. Jeyaprakash
- Ms. Anindita Rao
- Ms. K. Saraswathi
- Mr. Sethu Nagarajan
Senior Technician
- Mr. V. Saravanan
Technician
- Mr. D. Sathish Ponraj
Genetic Analyst
- Mr. S. Ulaganathan
Lab Assistants
- Mrs. D. Muthuselvi
- Ms. M.S. Indhumathi
ABI 3130 Genetic Analyzer

Genetic Analyzer has been installed in the Department of Molecular Genetics in 2007. Since then, it has been widely used by several groups of AMRF including Immunology, Microbiology and Proteomics. The sequencing instrument has been used for mutation screening, genotyping (Multi Locus Sequence Typing and Multiplex Ligation Dependent Probe Amplification), species/strain identification & confirmation and checking the clones. The data generated from sequencing in multiple research projects was presented in many national and international meetings and published in highly reputed journals of ophthalmology.
ABI 7900 HT Fast Real Time PCR

Real Time PCR has been installed in Department of Molecular Genetics in 2008. This machine has been used by Genetics, Microbiology and Ocular Pharmacology. This instrument is mainly used for the INDEYE study on genotyping Cataract and AMD samples by Allelic discrimination assay. The other research groups utilize this instrument for study of the gene expression, deletion/duplication analysis, copy number variations and also for species identification. So far more than six International research articles were published using the data generated from the instrument.